license: mit
IEPile: A Large-Scale Information Extraction Corpus
This is the official repository for IEPile: Unearthing Large-Scale Schema-Based Information Extraction Corpus
IEPile dataset download links: Google Drive | Hugging Face
We have meticulously collected and cleaned existing Information Extraction (IE) datasets, integrating a total of 26 English IE datasets and 7 Chinese IE datasets. As shown in Figure 1, these datasets cover multiple domains including general, medical, financial, and others.
In this study, we adopted the proposed "schema-based batched instruction generation method" to successfully create a large-scale, high-quality IE fine-tuning dataset named IEPile, containing approximately 0.32B tokens.
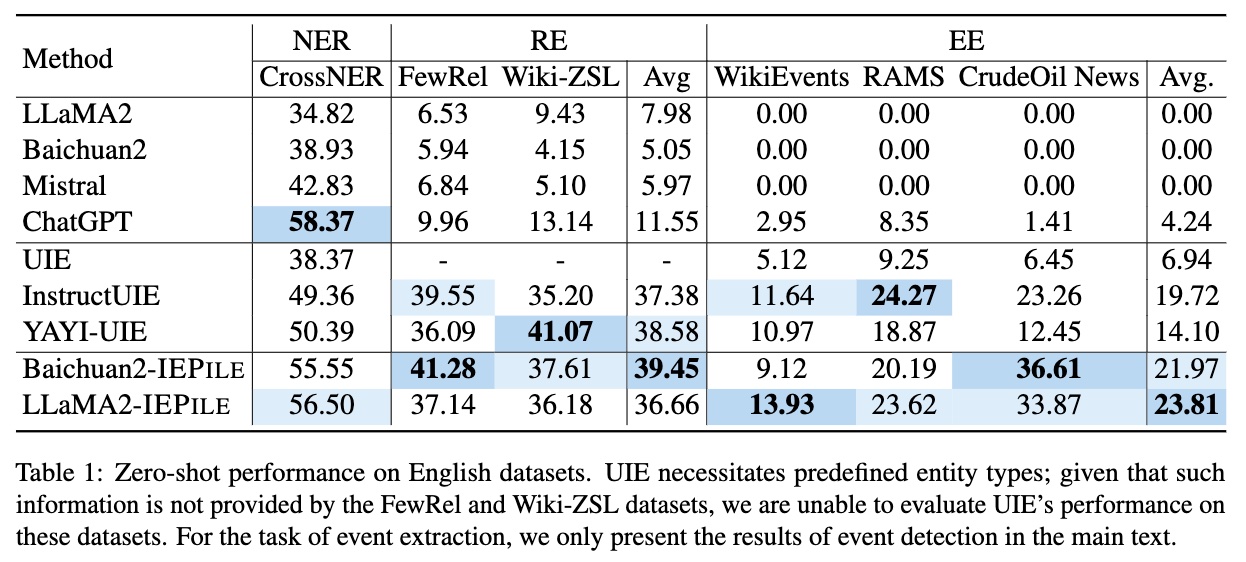
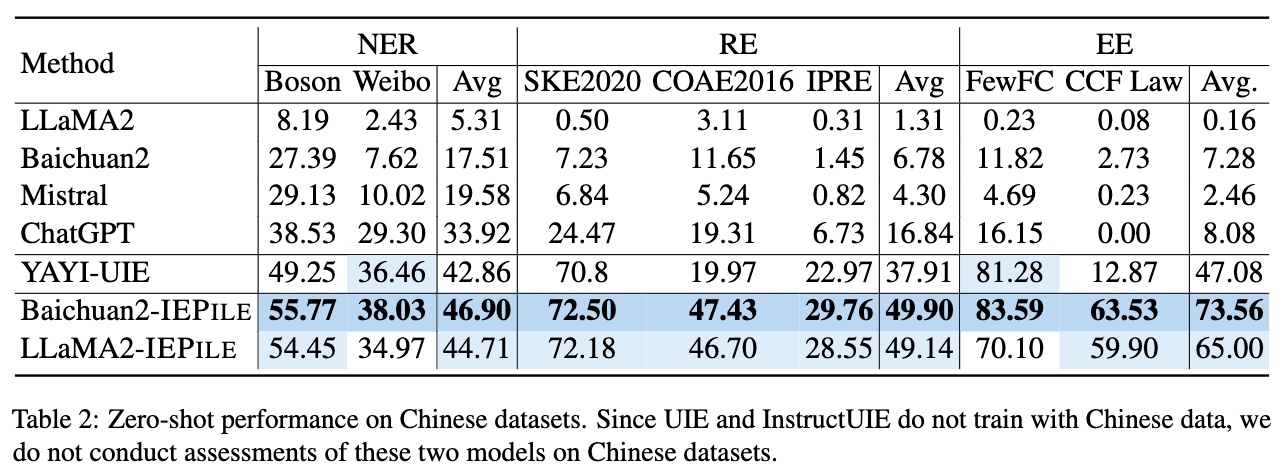
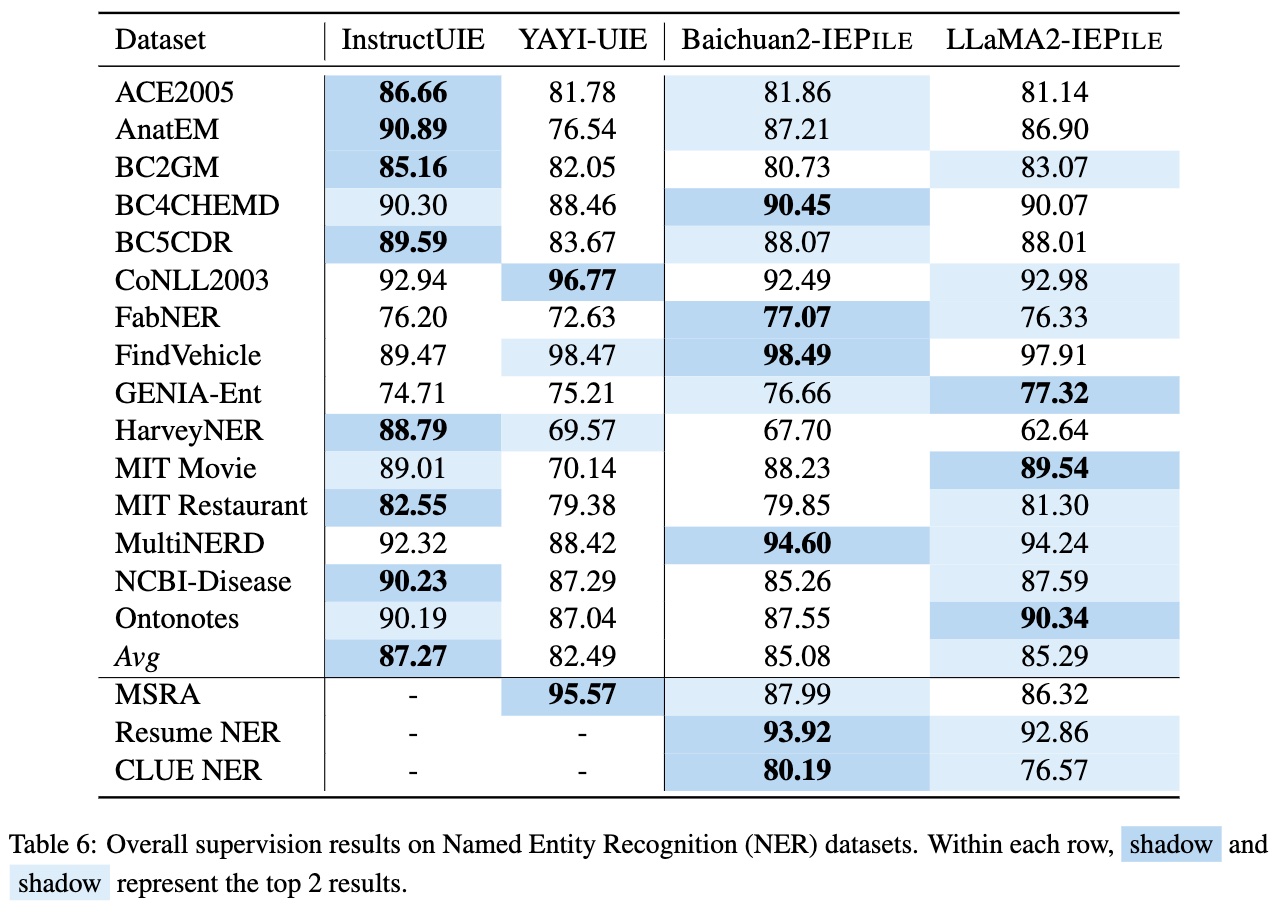
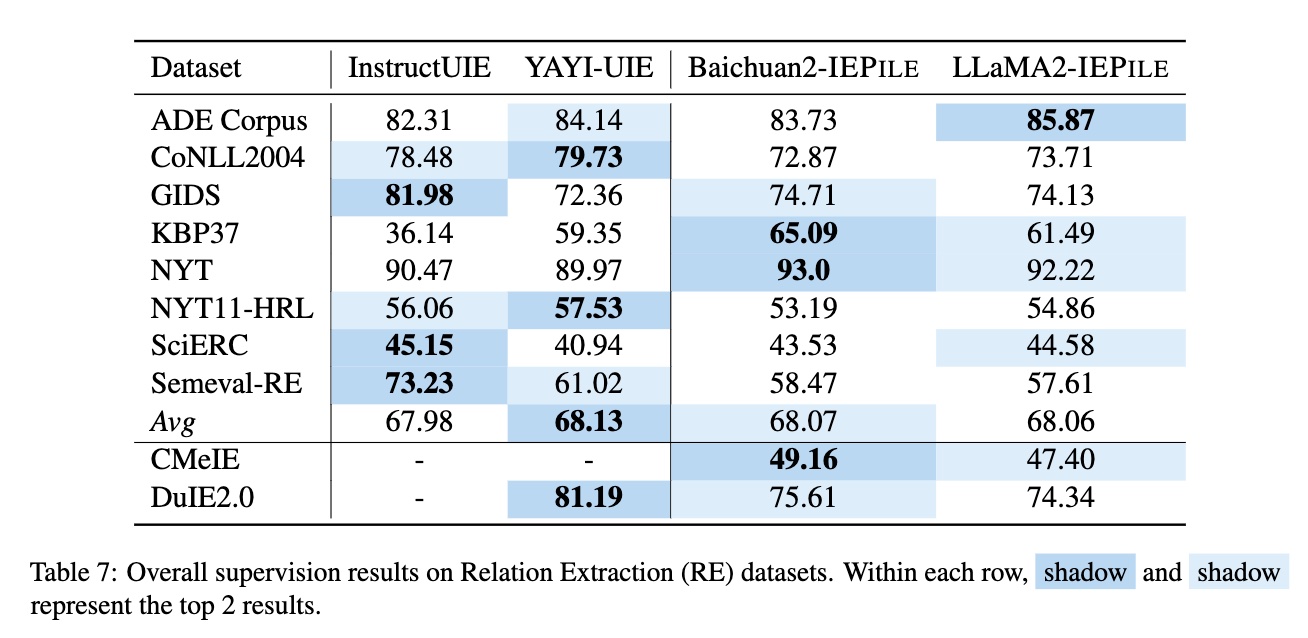
Based on IEPile, we fine-tuned the Baichuan2-13B-Chat and LLaMA2-13B-Chat models using the Lora technique. Experiments have demonstrated that the fine-tuned Baichuan2-IEPile and LLaMA2-IEPile models perform remarkably on fully supervised training sets and have achieved improvements in zero-shot information extraction tasks.
News
- [2024/02] We released a large-scale (0.32B tokens) high-quality bilingual (Chinese and English) Information Extraction (IE) instruction dataset named IEPile, along with two models trained on
IEPile, baichuan2-13b-iepile-lora and llama2-13b-iepile-lora. - [2023/10] We released a new bilingual (Chinese and English) theme-based Information Extraction (IE) instruction dataset named InstructIE with paper.
- [2023/08] We introduced a dedicated 13B model for Information Extraction (IE), named knowlm-13b-ie.
- [2023/05] We initiated an instruction-based Information Extraction project.
2.2Data Format
In IEPile, the instruction format of IEPile adopts a JSON-like string structure, which is essentially a dictionary-type string composed of the following three main components:
(1) 'instruction': Task description, which outlines the task to be performed by the instruction (one of NER, RE, EE, EET, EEA).
(2) 'schema': A list of schemas to be extracted (entity types, relation types, event types).
(3) 'input': The text from which information is to be extracted.
We recommend that you keep the number of schemas in each instruction to a fixed number, which is 6 for NER, and 4 for RE, EE, EET, EEA, as these are the quantities we used in our training.
instruction_mapper = {
'NERzh': "你是专门进行实体抽取的专家。请从input中抽取出符合schema定义的实体,不存在的实体类型返回空列表。请按照JSON字符串的格式回答。",
'REzh': "你是专门进行关系抽取的专家。请从input中抽取出符合schema定义的关系三元组,不存在的关系返回空列表。请按照JSON字符串的格式回答。",
'EEzh': "你是专门进行事件提取的专家。请从input中抽取出符合schema定义的事件,不存在的事件返回空列表,不存在的论元返回NAN,如果论元存在多值请返回列表。请按照JSON字符串的格式回答。",
'EETzh': "你是专门进行事件提取的专家。请从input中抽取出符合schema定义的事件类型及事件触发词,不存在的事件返回空列表。请按照JSON字符串的格式回答。",
'EEAzh': "你是专门进行事件论元提取的专家。请从input中抽取出符合schema定义的事件论元及论元角色,不存在的论元返回NAN或空字典,如果论元存在多值请返回列表。请按照JSON字符串的格式回答。",
'NERen': "You are an expert in named entity recognition. Please extract entities that match the schema definition from the input. Return an empty list if the entity type does not exist. Please respond in the format of a JSON string.",
'REen': "You are an expert in relationship extraction. Please extract relationship triples that match the schema definition from the input. Return an empty list for relationships that do not exist. Please respond in the format of a JSON string.",
'EEen': "You are an expert in event extraction. Please extract events from the input that conform to the schema definition. Return an empty list for events that do not exist, and return NAN for arguments that do not exist. If an argument has multiple values, please return a list. Respond in the format of a JSON string.",
'EETen': "You are an expert in event extraction. Please extract event types and event trigger words from the input that conform to the schema definition. Return an empty list for non-existent events. Please respond in the format of a JSON string.",
'EEAen': "You are an expert in event argument extraction. Please extract event arguments and their roles from the input that conform to the schema definition, which already includes event trigger words. If an argument does not exist, return NAN or an empty dictionary. Please respond in the format of a JSON string.",
}
split_num_mapper = {'NER':6, 'RE':4, 'EE':4, 'EET':4, 'EEA':4}
import json
task = 'NER'
language = 'en'
schema = ['person', 'organization', 'else', 'location']
split_num = split_num_mapper[task]
split_schemas = [schema[i:i+split_num] for i in range(0, len(schema), split_num)]
input = '284 Robert Allenby ( Australia ) 69 71 71 73 , Miguel Angel Martin ( Spain ) 75 70 71 68 ( Allenby won at first play-off hole )'
sintructs = []
for split_schema in split_schemas:
sintruct = json.dumps({'instruction':instruction_mapper[task+language], 'schema':split_schema, 'input':input}, ensure_ascii=False)
sintructs.append(sintruct)
More Tasks Schema
RE schema: ["neighborhood of", "nationality", "children", "place of death"] EE schema: [{"event_type": "potential therapeutic event", "trigger":True, "arguments": ["Treatment.Time_elapsed", "Treatment.Route", "Treatment.Freq", "Treatment", "Subject.Race", "Treatment.Disorder", "Effect", "Subject.Age", "Combination.Drug", "Treatment.Duration", "Subject.Population", "Subject.Disorder", "Treatment.Dosage", "Treatment.Drug"]}, {"event_type": "adverse event", "trigger":True, "arguments": ["Subject.Population", "Subject.Age", "Effect", "Treatment.Drug", "Treatment.Dosage", "Treatment.Freq", "Subject.Gender", "Treatment.Disorder", "Subject", "Treatment", "Treatment.Time_elapsed", "Treatment.Duration", "Subject.Disorder", "Subject.Race", "Combination.Drug"]}] EET schema: ["potential therapeutic event", "adverse event"] EEA schema: [{"event_type": "potential therapeutic event", "arguments": ["Treatment.Time_elapsed", "Treatment.Route", "Treatment.Freq", "Treatment", "Subject.Race", "Treatment.Disorder", "Effect", "Subject.Age", "Combination.Drug", "Treatment.Duration", "Subject.Population", "Subject.Disorder", "Treatment.Dosage", "Treatment.Drug"]}, {"event_type": "adverse event", "arguments": ["Subject.Population", "Subject.Age", "Effect", "Treatment.Drug", "Treatment.Dosage", "Treatment.Freq", "Subject.Gender", "Treatment.Disorder", "Subject", "Treatment", "Treatment.Time_elapsed", "Treatment.Duration", "Subject.Disorder", "Subject.Race", "Combination.Drug"]}]
Using baichuan2-13b-iepile-lora
import torch
from transformers import (
AutoConfig,
AutoTokenizer,
AutoModelForCausalLM,
GenerationConfig
)
from peft import PeftModel
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model_path = 'meta-llama/Llama-2-13b-chat-hf'
lora_path = 'zjunlp/llama2-13b-iepile-lora'
config = AutoConfig.from_pretrained(model_path, trust_remote_code=True)
tokenizer = AutoTokenizer.from_pretrained(model_path, trust_remote_code=True)
model = AutoModelForCausalLM.from_pretrained(
model_path,
config=config,
device_map="auto",
torch_dtype=torch.bfloat16,
trust_remote_code=True,
)
model = PeftModel.from_pretrained(
model,
lora_path,
)
model.eval()
system_prompt = "<<SYS>>\nYou are a helpful, respectful and honest assistant. Always answer as helpfully as possible, while being safe. Your answers should not include any harmful, unethical, racist, sexist, toxic, dangerous, or illegal content. Please ensure that your responses are socially unbiased and positive in nature.\n\nIf a question does not make any sense, or is not factually coherent, explain why instead of answering something not correct. If you don't know the answer to a question, please don't share false information.\n<</SYS>>\n\n"
sintruct = "{\"instruction\": \"You are an expert in named entity recognition. Please extract entities that match the schema definition from the input. Return an empty list if the entity type does not exist. Please respond in the format of a JSON string.\", \"schema\": [\"person\", \"organization\", \"else\", \"location\"], \"input\": \"284 Robert Allenby ( Australia ) 69 71 71 73 , Miguel Angel Martin ( Spain ) 75 70 71 68 ( Allenby won at first play-off hole )\"}"
sintruct = '[INST] ' + system_prompt + sintruct + ' [/INST]'
input_ids = tokenizer.encode(sintruct, return_tensors="pt").to(device)
input_length = input_ids.size(1)
generation_output = model.generate(input_ids=input_ids, generation_config=GenerationConfig(max_length=512, max_new_tokens=256, return_dict_in_generate=True))
generation_output = generation_output.sequences[0]
generation_output = generation_output[input_length:]
output = tokenizer.decode(generation_output, skip_special_tokens=True)
print(output)
If your GPU has limited memory, you can use quantization to reduce memory usage. Below is the inference process using 4-bit quantization.
import torch
from transformers import BitsAndBytesConfig
quantization_config=BitsAndBytesConfig(
load_in_4bit=True,
llm_int8_threshold=6.0,
llm_int8_has_fp16_weight=False,
bnb_4bit_compute_dtype=torch.bfloat16,
bnb_4bit_use_double_quant=True,
bnb_4bit_quant_type="nf4",
)
model = AutoModelForCausalLM.from_pretrained(
model_path,
config=config,
device_map="auto",
quantization_config=quantization_config,
torch_dtype=torch.bfloat16,
trust_remote_code=True,
)
model = PeftModel.from_pretrained(
model,
lora_path,
)
9.Cite
If you use the IEPile or the code, please cite the paper:
@article{DBLP:journals/corr/abs-2402-14710,
author = {Honghao Gui and
Lin Yuan and
Hongbin Ye and
Ningyu Zhang and
Mengshu Sun and
Lei Liang and
Huajun Chen},
title = {IEPile: Unearthing Large-Scale Schema-Based Information Extraction
Corpus},
journal = {CoRR},
volume = {abs/2402.14710},
year = {2024},
url = {https://doi.org/10.48550/arXiv.2402.14710},
doi = {10.48550/ARXIV.2402.14710},
eprinttype = {arXiv},
eprint = {2402.14710},
timestamp = {Tue, 09 Apr 2024 07:32:43 +0200},
biburl = {https://dblp.org/rec/journals/corr/abs-2402-14710.bib},
bibsource = {dblp computer science bibliography, https://dblp.org}
}