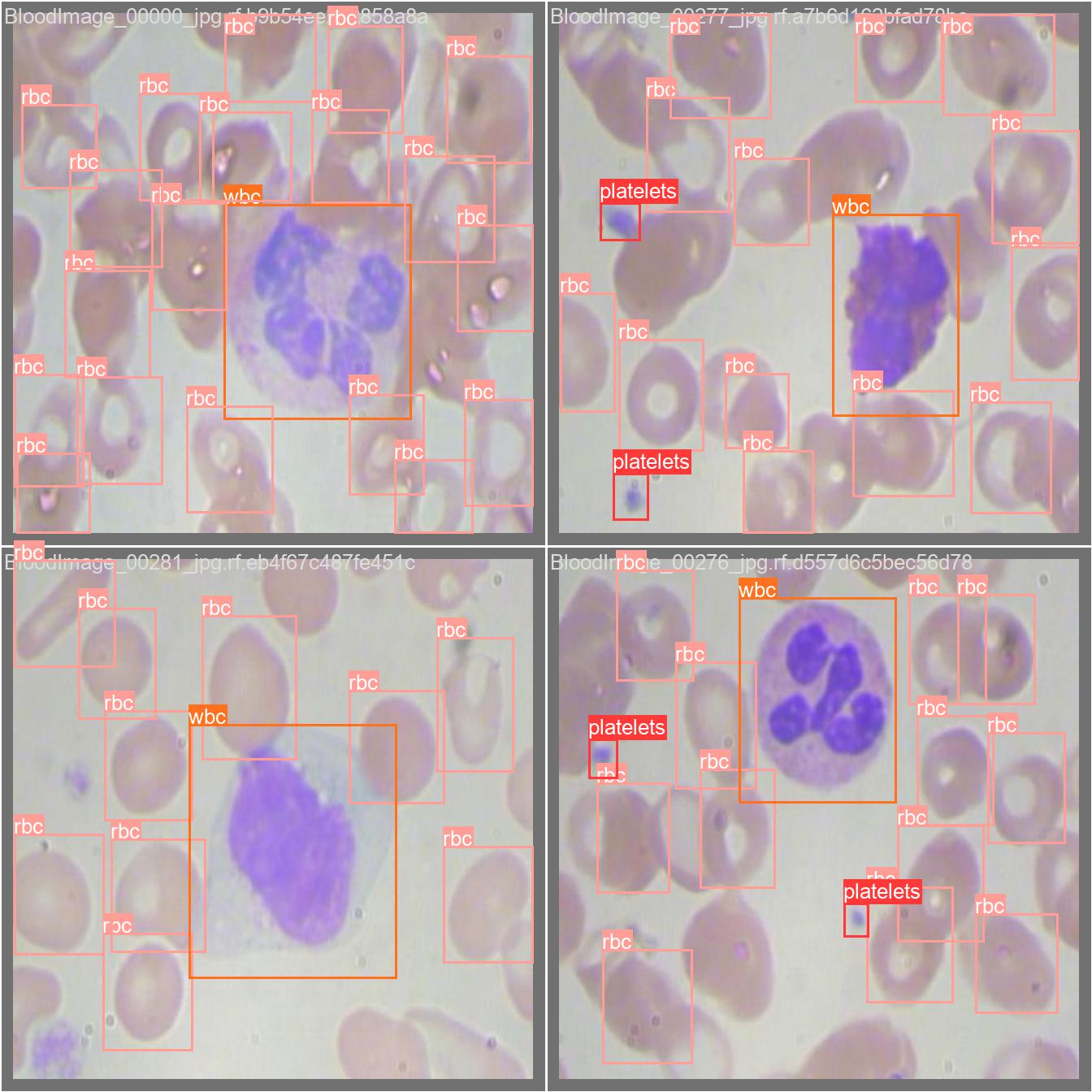
How to use
- Install yolov5:
pip install -U yolov5
- Load model and perform prediction:
import yolov5
# load model
model = yolov5.load('keremberke/yolov5m-blood-cell')
# set model parameters
model.conf = 0.25 # NMS confidence threshold
model.iou = 0.45 # NMS IoU threshold
model.agnostic = False # NMS class-agnostic
model.multi_label = False # NMS multiple labels per box
model.max_det = 1000 # maximum number of detections per image
# set image
img = 'https://github.com/ultralytics/yolov5/raw/master/data/images/zidane.jpg'
# perform inference
results = model(img, size=640)
# inference with test time augmentation
results = model(img, augment=True)
# parse results
predictions = results.pred[0]
boxes = predictions[:, :4] # x1, y1, x2, y2
scores = predictions[:, 4]
categories = predictions[:, 5]
# show detection bounding boxes on image
results.show()
# save results into "results/" folder
results.save(save_dir='results/')
- Finetune the model on your custom dataset:
yolov5 train --data data.yaml --img 640 --batch 16 --weights keremberke/yolov5m-blood-cell --epochs 10
More models available at: awesome-yolov5-models
- Downloads last month
- 869
Dataset used to train keremberke/yolov5m-blood-cell
Space using keremberke/yolov5m-blood-cell 1
Evaluation results
- [email protected] on keremberke/blood-cell-object-detectionvalidation set self-reported0.905